Hacohen Lab
The Hacohen lab consists of immunologists, geneticists, biochemists, technologists and computational biologists working together to understand basic immune processes and immune-mediated diseases.

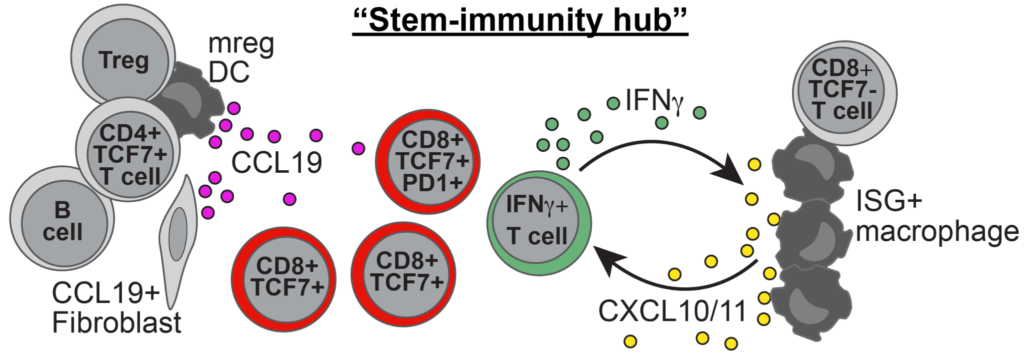
The organization of immune cells in human tumors is not well understood. Immunogenic tumors harbor spatially localized multicellular ‘immunity hubs’ defined by expression of the T cell-attracting chemokines CXCL10/CXCL11 and abundant T cells. Here, we examined immunity hubs in human pre-immunotherapy lung cancer specimens and found an association with beneficial response to PD-1 blockade. Critically, we discovered the stem-immunity hub, a subtype of immunity hub strongly associated with favorable PD-1-blockade outcome. This hub is distinct from mature tertiary lymphoid structures and is enriched for stem-like TCF7+PD-1+CD8+ T cells, activated CCR7+LAMP3+ dendritic cells and CCL19+ fibroblasts as well as chemokines that organize these cells. Within the stem-immunity hub, we find preferential interactions between CXCL10+ macrophages and TCF7−CD8+ T cells as well as between mature regulatory dendritic cells and TCF7+CD4+ and regulatory T cells. These results provide a picture of the spatial organization of the human intratumoral immune response and its relevance to patient immunotherapy outcomes.
pH measurement in cells
| unstimulated | STING agonist | STING agonist with channel blocker |
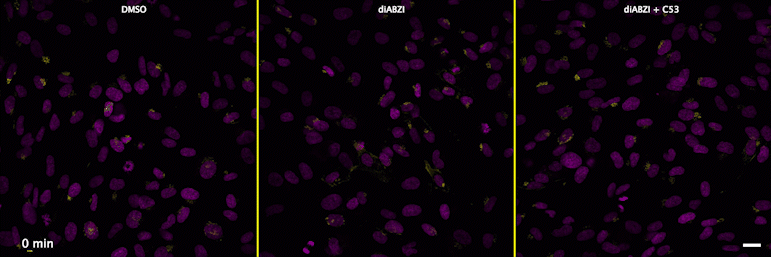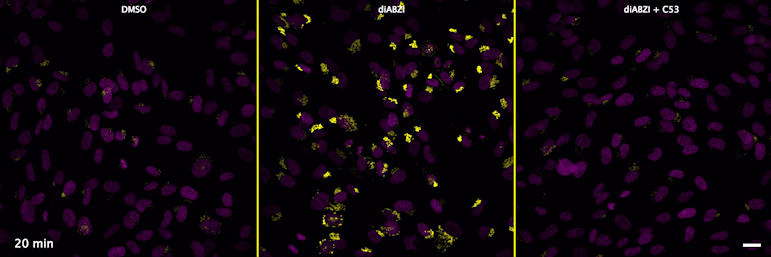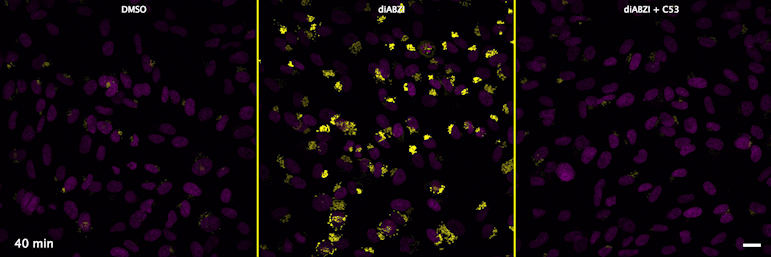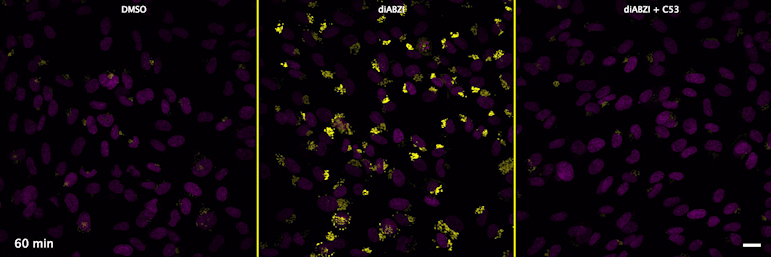
Human STING is a proton channel (2023) (link to Science here).
Through a convergence of several concepts, Bingxu Liu, a graduate student in our lab and Darrell Irvine’s lab, hypothesized that human STING is a proton channel that could potentially activate key cellular processes. To test this hypothesis, Bingxu, Becca Carlson and Ivan Pires carried out experiments in cells and in vitro and found that: (1) STING activation induced a pH increase in the Golgi; (2) STING reconstituted in liposomes enabled transmembrane proton transport; (3) the small molecule C53 (a STING agonist that binds the putative channel interface based on published structures) blocked STING-induced proton flux in the Golgi and in liposomes, and inhibited LC3B lipidation and inflammasome activation in cells. Sting’s role as channel is thus to trigger LC3B lipidation and inflammasome activation, processes which appear decoupled from STING’s interferon-inducing function.
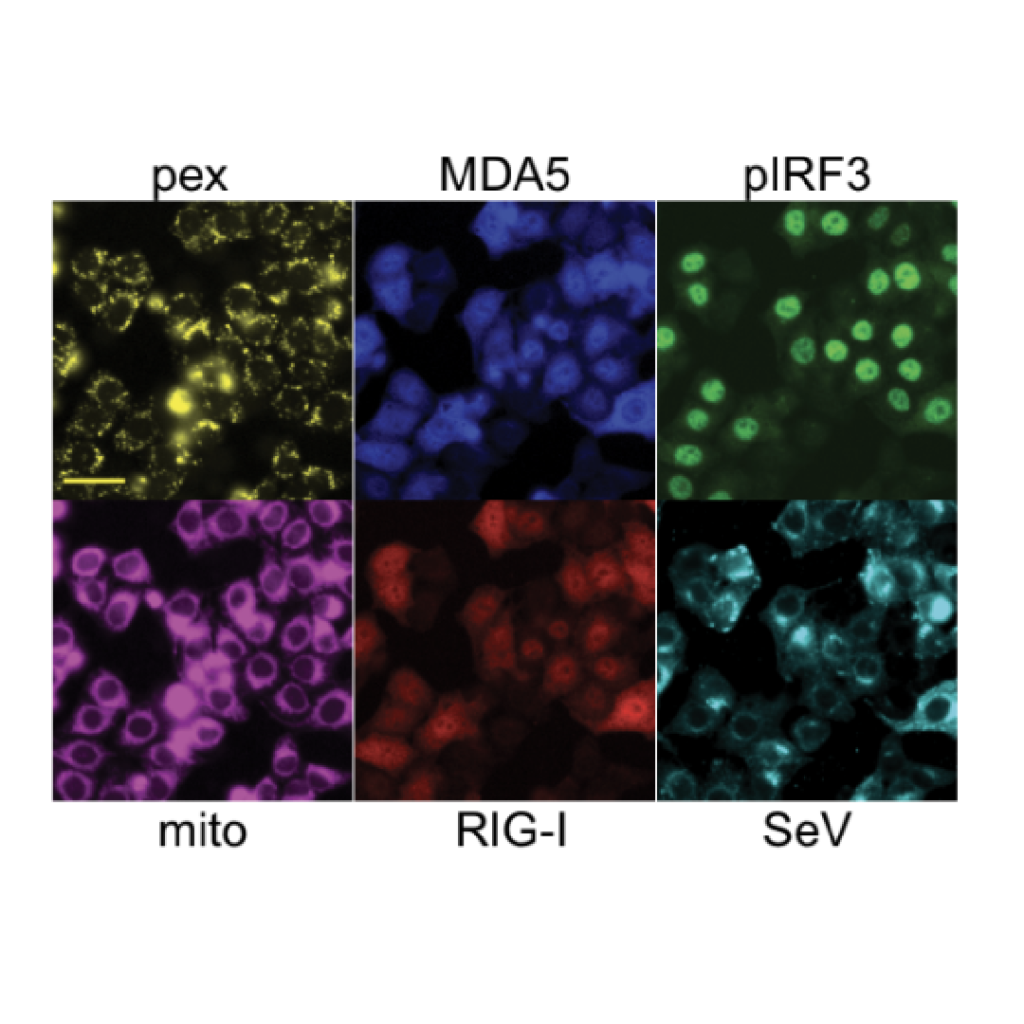
A genome-wide optical pooled screen reveals regulators of cellular antiviral responses (2023)
Optical pooled screens pair single-cell phenotypic imaging with large-scale CRISPR or other perturbations, providing rich information about complex processes in huge numbers of cells at a time. Rebecca Carlson, Paul Blainey and our collaborators have expanded the scope of optical pooled screens genome-wide and deployed the technology to explore how cells regulate their responses to RNA virus infection. The genome-wide screens revealed previously unrecognized aspects of viral sensing in cells, and demonstrated how conventional and machine learning-enhanced computer vision-based phenotyping and genome-wide pooled CRISPR screening expand the scope of functional genomics research.
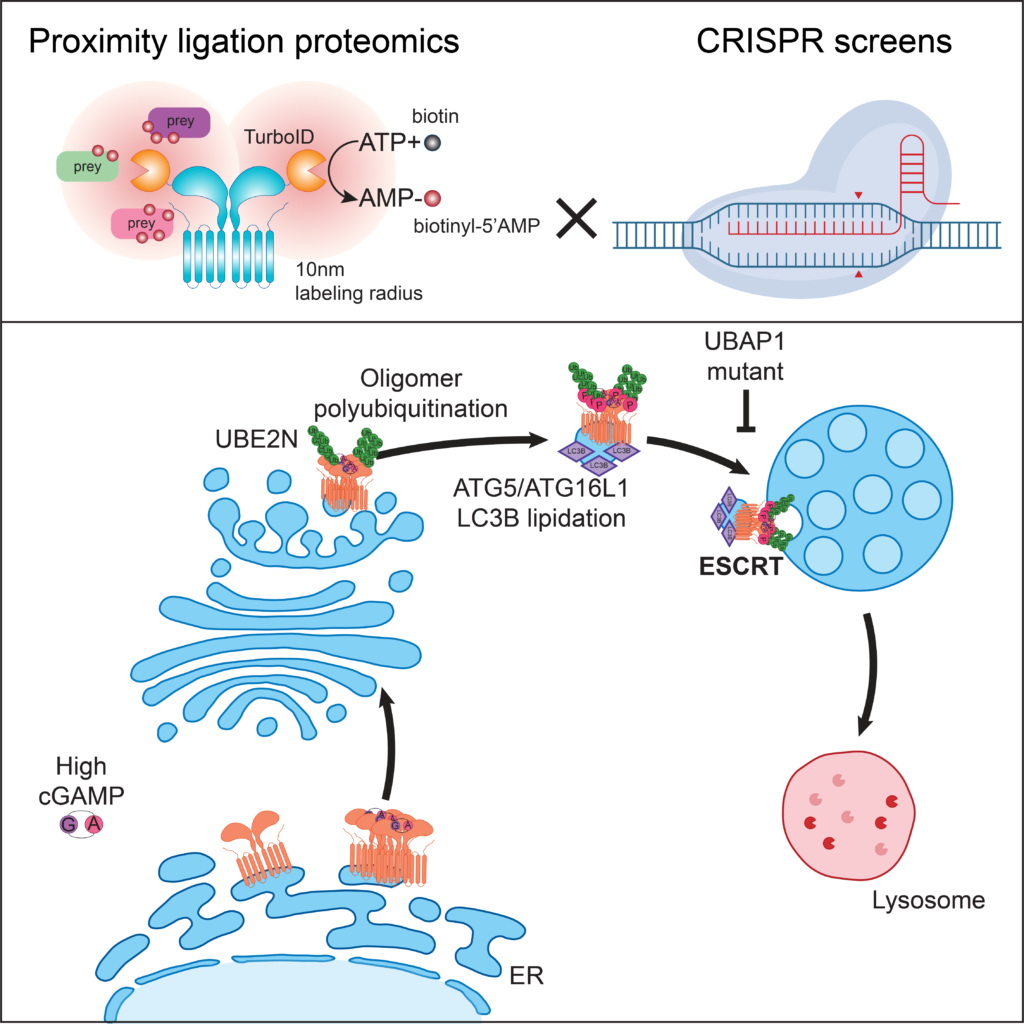
ESCRT-dependent STING degradation inhibits steady-state and cGAMP-induced signalling (2022)
Systems approaches to elucidate genes regulating STING trafficking. By combining proximity ligation proteomics and CRISPR screens, Matteo Gentili, a pdoc in the lab, showed that the ESCRT complex is required for STING degradation at the lysosome. ESCRT regulates a homeostatic STING degradative flux and an ESCRT mutant identified in patients with hereditary spastic paraplegia increases steady-state STING-dependent type I IFN responses.