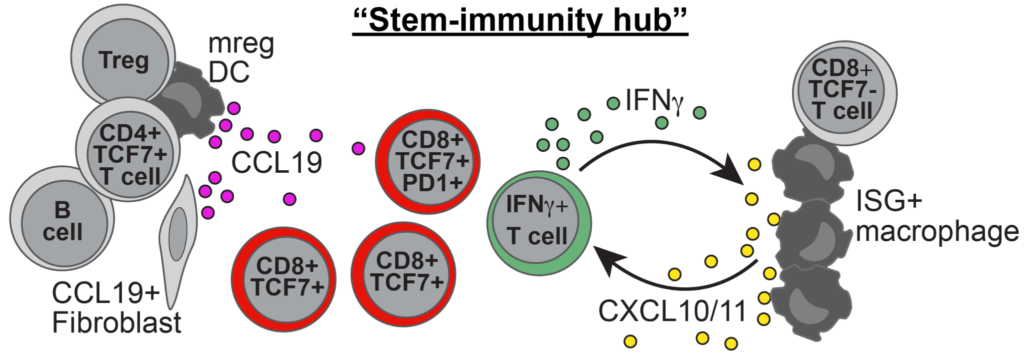
Human lung cancer harbors spatially organized stem-immunity hubs associated with response to immunotherapy (2024)
The organization of immune cells in human tumors is not well understood. Immunogenic tumors harbor spatially localized multicellular ‘immunity hubs’ defined by expression of the T cell-attracting chemokines CXCL10/CXCL11 and abundant T cells. Here, we examined immunity hubs in human pre-immunotherapy lung cancer specimens and found an association with beneficial response to PD-1 blockade. Critically, we discovered the stem-immunity hub, a subtype of immunity hub strongly associated with favorable PD-1-blockade outcome. This hub is distinct from mature tertiary lymphoid structures and is enriched for stem-like TCF7+PD-1+CD8+ T cells, activated CCR7+LAMP3+ dendritic cells and CCL19+ fibroblasts as well as chemokines that organize these cells. Within the stem-immunity hub, we find preferential interactions between CXCL10+ macrophages and TCF7−CD8+ T cells as well as between mature regulatory dendritic cells and TCF7+CD4+ and regulatory T cells. These results provide a picture of the spatial organization of the human intratumoral immune response and its relevance to patient immunotherapy outcomes.
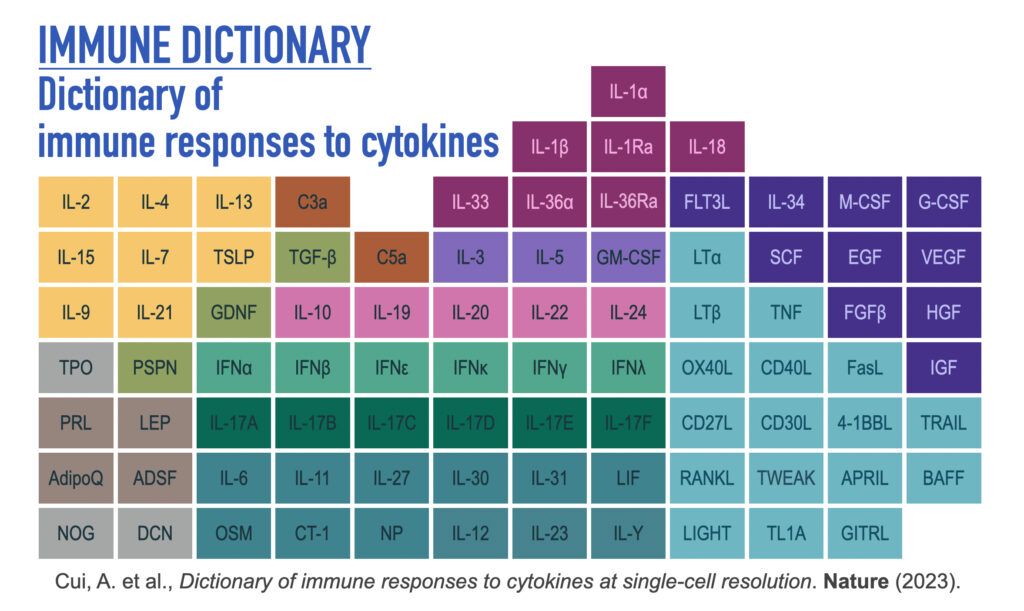
Dictionary of immune responses to cytokines at single-cell resolution (2023)
Cytokines mediate cell–cell communication in the immune system and represent important therapeutic targets1,2,3. A myriad of studies have highlighted their central role in immune function4,5,6,7,8,9,10,11,12,13, yet we lack a global view of the cellular responses of each immune cell type to each cytokine. To address this gap, we created the Immune Dictionary, a compendium of single-cell transcriptomic profiles of more than 17 immune cell types in response to each of 86 cytokines (>1,400 cytokine–cell type combinations) in mouse lymph nodes in vivo. A cytokine-centric view of the dictionary revealed that most cytokines induce highly cell-type-specific responses. For example, the inflammatory cytokine interleukin-1β induces distinct gene programmes in almost every cell type. A cell-type-centric view of the dictionary identified more than 66 cytokine-driven cellular polarization states across immune cell types, including previously uncharacterized states such as an interleukin-18-induced polyfunctional natural killer cell state. Based on this dictionary, we developed companion software, Immune Response Enrichment Analysis, for assessing cytokine activities and immune cell polarization from gene expression data, and applied it to reveal cytokine networks in tumours following immune checkpoint blockade therapy. Our dictionary generates new hypotheses for cytokine functions, illuminates pleiotropic effects of cytokines, expands our knowledge of activation states of each immune cell type, and provides a framework to deduce the roles of specific cytokines and cell–cell communication networks in any immune response. You can download the dataset here.
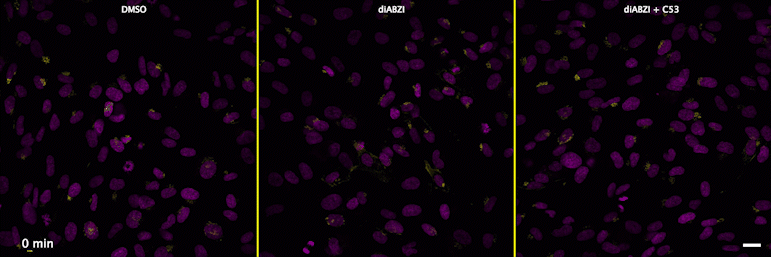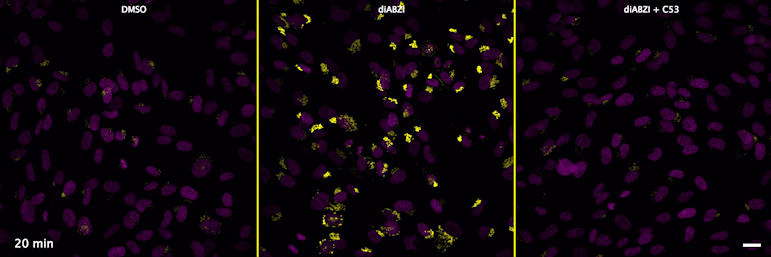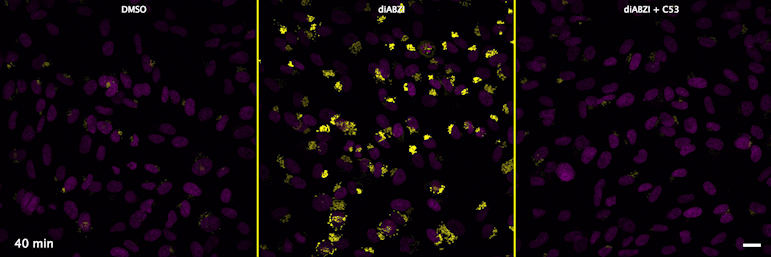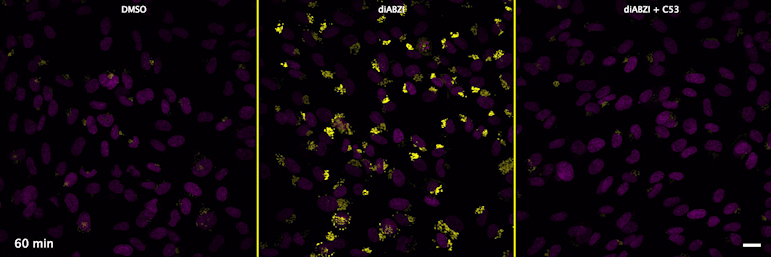
Human STING is a proton channel (2023)
Through a convergence of several concepts, Bingxu Liu, a graduate student in our lab and Darrell Irvine’s lab, hypothesized that human STING is a proton channel that could potentially activate key cellular processes. To test this hypothesis, Bingxu, Becca Carlson and Ivan Pires carried out experiments in cells and in vitro and found that: (1) STING activation induced a pH increase in the Golgi; (2) STING reconstituted in liposomes enabled transmembrane proton transport; (3) the small molecule C53 (a STING agonist that binds the putative channel interface based on published structures) blocked STING-induced proton flux in the Golgi and in liposomes, and inhibited LC3B lipidation and inflammasome activation in cells. Sting’s role as channel is thus to trigger LC3B lipidation and inflammasome activation, processes which appear decoupled from STING’s interferon-inducing function.
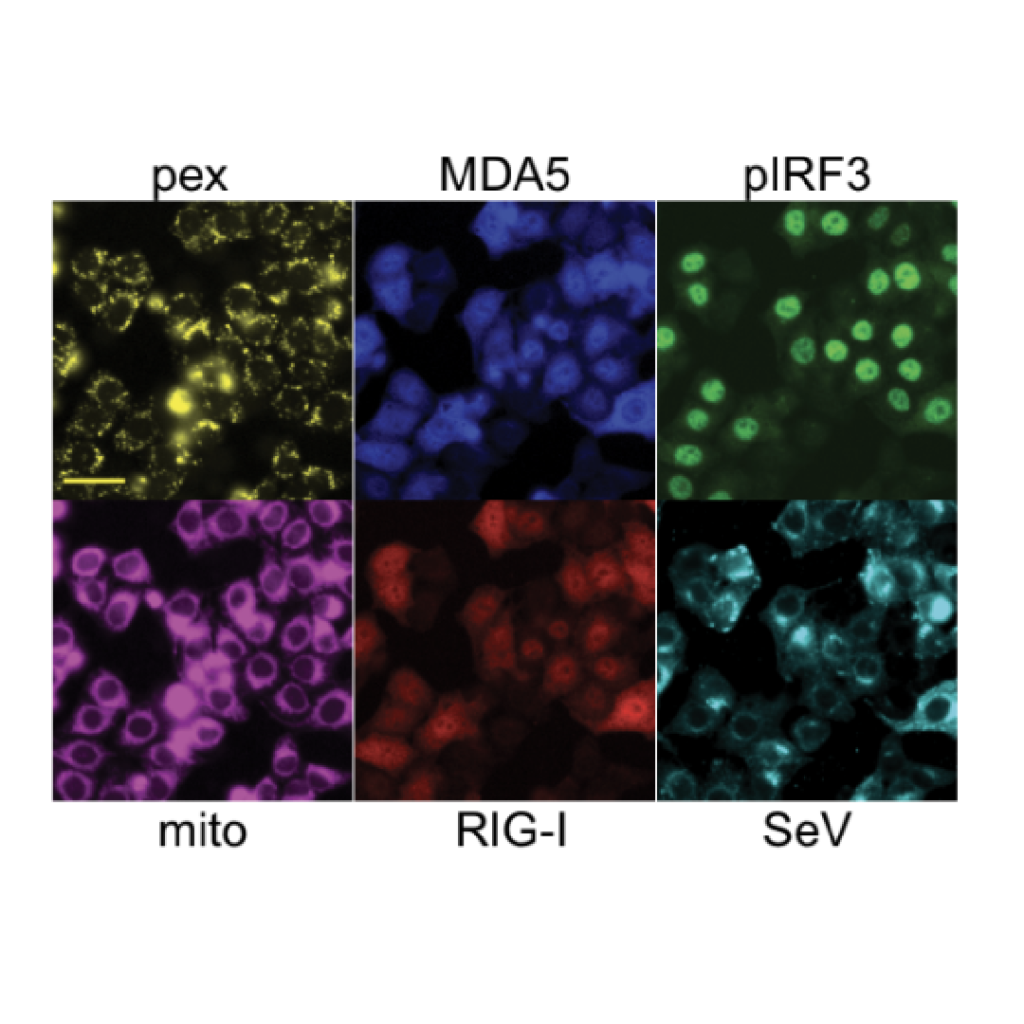
A genome-wide optical pooled screen reveals regulators of cellular antiviral responses (2023)
Optical pooled screens pair single-cell phenotypic imaging with large-scale CRISPR or other perturbations, providing rich information about complex processes in huge numbers of cells at a time. Rebecca Carlson, Paul Blainey and our collaborators have expanded the scope of optical pooled screens genome-wide and deployed the technology to explore how cells regulate their responses to RNA virus infection. The genome-wide screens revealed previously unrecognized aspects of viral sensing in cells, and demonstrated how conventional and machine learning-enhanced computer vision-based phenotyping and genome-wide pooled CRISPR screening expand the scope of functional genomics research.
Combined PD-1, BRAF and MEK inhibition in BRAFV600E colorectal cancer: a phase 2 trial. (2022)
In a successful clinical trial for BRAF mutant colorectal cancer, we used single cell RNA sequencing of tumor epithelial cells from biopsies to show that combining BRAF/MEK inhibition with PD-1 blockade resulted in not only inhibition of the MAPK pathway but also increased expression of interferon stimulated genes in patients who benefitted from the treatment.
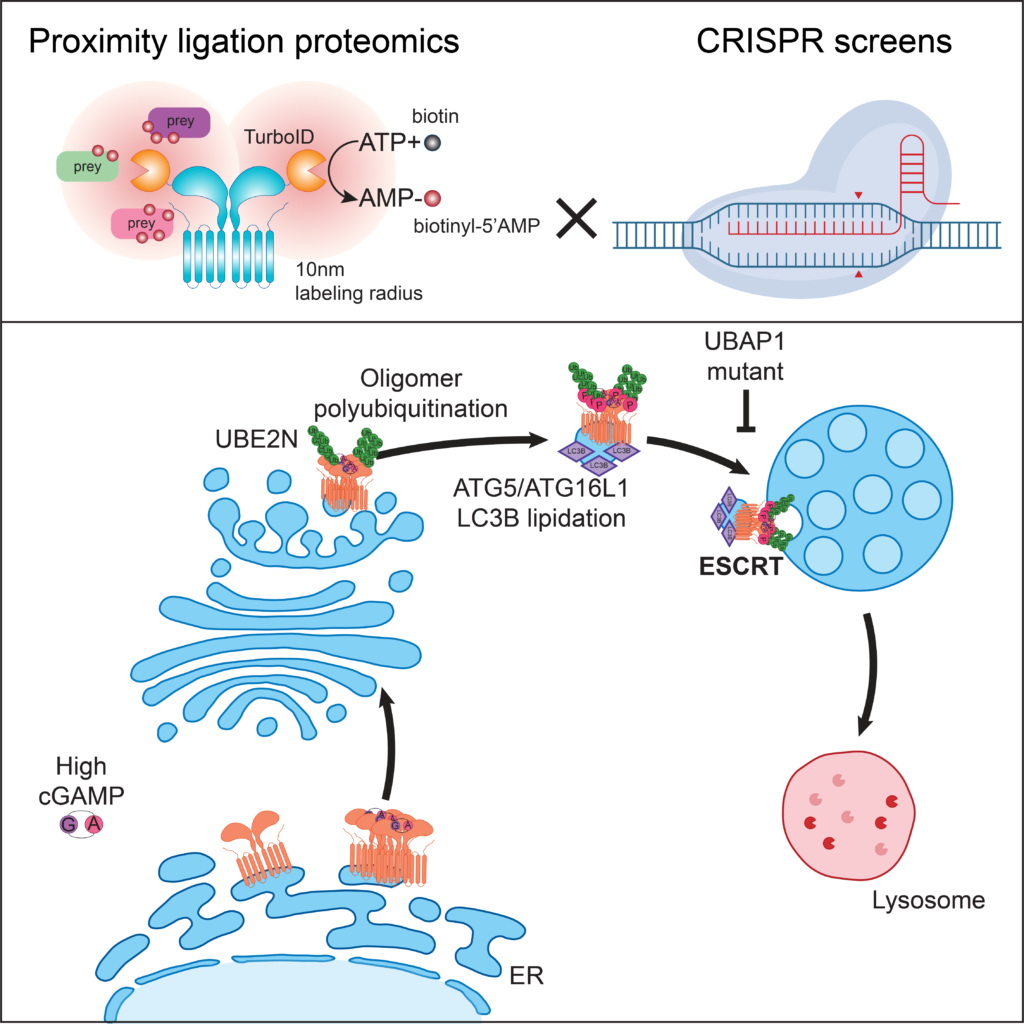
ESCRT-dependent STING degradation inhibits steady-state and cGAMP-induced signalling. (2022)
Systems approaches to elucidate genes regulating STING trafficking. By combining proximity ligation proteomics and CRISPR screens, Matteo Gentili, a pdoc in the lab, showed that the ESCRT complex is required for STING degradation at the lysosome. ESCRT regulates a homeostatic STING degradative flux and an ESCRT mutant identified in patients with hereditary spastic paraplegia increases steady-state STING-dependent type I IFN responses.
Selected Publications
Cui, A., Huang, T., Li, S. et al. Dictionary of immune responses to cytokines at single-cell resolution. Nature (2023). https://doi.org/10.1038/s41586-023-06816-9
Liu B, Carlson RJ, Pires IS, Gentili M, Feng E, Hellier Q, Schwartz MA, Blainey PC, Irvine DJ, Hacohen N. Human STING is a proton channel. Science. 2023 Aug 4;381(6657):508-514. doi: 10.1126/science.adf8974. Epub 2023 Aug 3. PMID: 37535724.
Tian J, Chen JH, Chao SX, Pelka K, Giannakis M, Hess J, Burke K, Jorgji V, Sindurakar P, Braverman J, Mehta A, Oka T, Huang M, Lieb D, Spurrell M, Allen JN, Abrams TA, Clark JW, Enzinger AC, Enzinger PC, Klempner SJ, McCleary NJ, Meyerhardt JA, Ryan DP, Yurgelun MB, Kanter K, Van Seventer EE, Baiev I, Chi G, Jarnagin J, Bradford WB, Wong E, Michel AG, Fetter IJ, Siravegna G, Gemma AJ, Sharpe A, Demehri S, Leary R, Campbell CD, Yilmaz O, Getz GA, Parikh AR, Hacohen N, Corcoran RB. Combined PD-1, BRAF and MEK inhibition in BRAFV600E colorectal cancer: a phase 2 trial. Nat Med. 2023 Feb;29(2):458-466. doi: 10.1038/s41591-022-02181-8. Epub 2023 Jan 26. PMID: 36702949; PMCID: PMC9941044
Al’Khafaji AM, Smith JT, Garimella KV, Babadi M, Popic V, Sade-Feldman M, Gatzen M, Sarkizova S, Schwartz MA, Blaum EM, Day A, Costello M, Bowers T, Gabriel S, Banks E, Philippakis AA, Boland GM, Blainey PC, Hacohen N. High-throughput RNA isoform sequencing using programmed cDNA concatenation. Nat Biotechnol. 2023 Jun 8. doi: 10.1038/s41587-023-01815-7. PMID: 37291427.
Carlson RJ, Leiken MD, Guna A, Hacohen N, Blainey P. A genome-wide optical pooled screen reveals regulators of cellular antiviral responses. PNAS. 2023 Apr 12. 120 (16) e2210623120. doi:10.1073/pnas.2210623120.
Reyes M, Leff SM, Gentili M, Hacohen N, Blainey PC. Microscale combinatorial stimulation of human myeloid cells reveals inflammatory priming by viral ligands. Sci Adv. 2023 Feb 24;9(8):eade5090. doi: 10.1126/sciadv.ade5090. Epub 2023 Feb 24. PMID: 36827376; PMCID: PMC9956118.
Gentili M, Liu B, Papanastasiou M, Dele-Oni D, Schwartz MA, Carlson RJ, Al’Khafaji AM, Krug K, Brown A, Doench JG, Carr SA, Hacohen N. ESCRT-dependent STING degradation inhibits steady-state and cGAMP-induced signalling. Nat Commun. 2023 Feb 4;14(1):611. doi: 10.1038/s41467-023-36132-9. PMID: 36739287; PMCID: PMC9899276.
LaSalle TJ, Gonye ALK, Freeman SS, Kaplonek P, Gushterova I, Kays KR, Manakongtreecheep K, Tantivit J, Rojas-Lopez M, Russo BC, Sharma N, Thomas MF, Lavin-Parsons KM, Lilly BM, Mckaig BN, Charland NC, Khanna HK, Lodenstein CL, Margolin JD, Blaum EM, Lirofonis PB, Revach OY, Mehta A, Sonny A, Bhattacharyya RP, Parry BA, Goldberg MB, Alter G, Filbin MR, Villani AC, Hacohen N, Sade-Feldman M. Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients. Cell Rep Med. 2022 Oct 18;3(10):100779. doi: 10.1016/j.xcrm.2022.100779. Epub 2022 Sep 26. PMID: 36208629; PMCID: PMC9510054.
Mouri K, Guo MH, de Boer CG, Lissner MM, Harten IA, Newby GA, DeBerg HA, Platt WF, Gentili M, Liu DR, Campbell DJ, Hacohen N, Tewhey R, Ray JP. Prioritization of autoimmune disease-associated genetic variants that perturb regulatory element activity in T cells. Nat Genet. 2022 May;54(5):603-612. doi: 10.1038/s41588-022-01056-5. Epub 2022 May 5. PMID: 35513721; PMCID: PMC9793778.
Freeman SS, Sade-Feldman M, Kim J, Stewart C, Gonye ALK, Ravi A, Arniella MB, Gushterova I, LaSalle TJ, Blaum EM, Yizhak K, Frederick DT, Sharova T, Leshchiner I, Elagina L, Spiro OG, Livitz D, Rosebrock D, Aguet F, Carrot-Zhang J, Ha G, Lin Z, Chen JH, Barzily-Rokni M, Hammond MR, Vitzthum von Eckstaedt HC, Blackmon SM, Jiao YJ, Gabriel S, Lawrence DP, Duncan LM, Stemmer-Rachamimov AO, Wargo JA, Flaherty KT, Sullivan RJ, Boland GM, Meyerson M, Getz G, Hacohen N. Combined tumor and immune signals from genomes or transcriptomes predict outcomes of checkpoint inhibition in melanoma. Cell Rep Med. 2022 Feb 15;3(2):100500. doi: 10.1016/j.xcrm.2021.100500. PMID: 35243413; PMCID: PMC8861826.
Chao S, Brenner MP, Hacohen N. Identifying Cell Type-Specific Chemokine Correlates with Hierarchical Signal Extraction from Single-Cell Transcriptomes. Pac Symp Biocomput. 2022;27:254-265. PMID: 34890154.
Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V,…,Ng K, Giannakis M, Nieman LT, Boland GM, Aguirre AJ, Anderson AC, Rozenblatt-Rosen O, Regev A, Hacohen N. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021 Aug 24:S0092-8674(21)00945-4.
Reyes M, Filbin MR, Bhattacharyya RP, Sonny A, Mehta A, Billman K… Baron RM, Goldberg MB, Blainey PC, Hacohen N. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci Transl Med. 2021 Jun 16;13(598):eabe9599.
Reyes M, Filbin MR, Bhattacharyya RP, Billman K, Eisenhaure T, Hung DT, Levy BD, Baron RM, Blainey PC, Goldberg MB, Hacohen N. An immune-cell signature of bacterial sepsis. Nat Med. 2020 Mar;26(3):333-340. doi: 10.1038/s41591-020-0752-4. Epub 2020 Feb 17. PMID: 32066974; PMCID: PMC7235950.
Li B, Clohisey SM, Chia BS, Wang B, Cui A, Eisenhaure T, Schweitzer LD, Hoover P, Parkinson NJ, Nachshon A, Smith N, Regan T, Farr D, Gutmann MU, Bukhari SI, Law A, Sangesland M, Gat-Viks I, Digard P, Vasudevan S, Lingwood D, Dockrell DH, Doench JG, Baillie JK, Hacohen N. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat Commun. 2020 Jan 9;11(1):164. doi: 10.1038/s41467-019-13965-x. PMID: 31919360; PMCID: PMC6952391.
Sarkizova S, Klaeger S, Le PM, Li LW, Oliveira G, Keshishian H, Hartigan CR, Zhang W, Braun DA, Ligon KL, Bachireddy P, Zervantonakis IK, Rosenbluth JM, Ouspenskaia T, Law T, Justesen S, Stevens J, Lane WJ, Eisenhaure T, Lan Zhang G, Clauser KR, Hacohen N, Carr SA, Wu CJ, Keskin DB. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol. 2020 Feb;38(2):199-209. doi: 10.1038/s41587-019-0322-9.
Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, Diamond B; Accelerating Medicines Partnership in SLE network. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019 Jul;20(7):902-914. doi: 10.1038/s41590-019-0398-x. Epub 2019 Jun 17. Erratum in: Nat Immunol. 2019 Aug 13;: PMID: 31209404; PMCID: PMC6726437.
Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, Freeman SS, Reuben A, Hoover PJ, Villani A-C, Ivanova E, Portell A, Lizotte PH, Aref AR, Eliane JP, Hammond MR, Vitzthum H, Blackmon SM, Li B, Gopalakrishnan V, Reddy SM, Cooper ZA, Paweletz CP, Barbie DA, Stemmer- Rachamimov S, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G and Hacohen N. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018 Nov 1;175(4):998-1013.e20.
Ott P, Hu X, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, Chen C, Olive O, Carter TA, Li S, Lieb DJ, Eisenhaure T, Gjini E, Stevens J, Lane WJ, Javeri I, Nellaiappan K, Andreas Salazar12, Daley H, Seaman M, Buchbinder EI, Yoo CH, Harden M, Lennon N, Gabriel S, Rodig SJ, Barouch DH, Aster JC, Getz G, Wucherpfennig K, Neuberg D, Ritz J, Lander ES, Fritsch EF, Hacohen N & Wu CJ. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, Jul 13;547(7662):217-221.
Villani A-C, Satija R, Reynolds G, Shekhar K, Fletcher J, Sarkizova S, Griesbeck M, Butler A, Zheng S,Lazo S, Jardine L, Dixon D, Stephenson E, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes and progenitors. Science 2017, Apr 21;356(6335).
Jovanovic M, Rooney MS, Mertins P, Przybylski D, Chevrier N, Satija R, Rodriguez EH, Fields AP, Schwartz S, Raychowdhury R, Mumbach MR, Eisenhaure T, Rabani M, Gennert D, Lu D, Delorey T, Weissman JS, Carr SA, Hacohen N, Regev A. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015 Mar 6;347(6226):1259038. doi: 10.1126/science.1259038. Epub 2015 Feb 12. PMID: 25745177; PMCID: PMC4506746.
Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, Shalem O, Satija R, Raychowdhury R, Mertins P, Carr SA, Zhang F, Hacohen N, Regev A. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015 Jul 30;162(3):675-86. doi: 10.1016/j.cell.2015.06.059. Epub 2015 Jul 16. PMID: 26189680; PMCID: PMC4522370.
Lan YY, Londoño D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014 Oct 9;9(1):180-192. doi: 10.1016/j.celrep.2014.08.074. Epub 2014 Oct 2. PMID: 25284779; PMCID: PMC4555847.
Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, Ward LD, Raddassi K, McCabe C, Lee MH, Frohlich IY, Hafler DA, Kellis M, Raychaudhuri S, Zhang F, Stranger BE, Benoist CO, De Jager PL, Regev A, Hacohen N. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014 Mar 7;343(6175):1246980. doi: 10.1126/science.1246980. PMID: 24604203; PMCID: PMC4124741.
See All Publications